Note
Click here to download the full example code
Advanced processing
The pynapple package provides a small set of high-level functions that are widely used in systems neuroscience.
- Discrete correlograms
- Tuning curves
- Decoding
- PETH
- Randomization
This notebook provides few examples with artificial data.
Warning
This tutorial uses seaborn and matplotlib for displaying the figure.
You can install both with pip install matplotlib seaborn
import numpy as np
import pandas as pd
import pynapple as nap
import matplotlib.pyplot as plt
import seaborn as sns
custom_params = {"axes.spines.right": False, "axes.spines.top": False}
sns.set_theme(style="ticks", palette="colorblind", font_scale=1.5, rc=custom_params)
Discrete correlograms
First let's generate some data. Here we have two neurons recorded together. We can group them in a TsGroup.
ts1 = nap.Ts(t=np.sort(np.random.uniform(0, 1000, 2000)), time_units="s")
ts2 = nap.Ts(t=np.sort(np.random.uniform(0, 1000, 1000)), time_units="s")
epoch = nap.IntervalSet(start=0, end=1000, time_units="s")
ts_group = nap.TsGroup({0: ts1, 1: ts2}, time_support=epoch)
print(ts_group)
Out:
First we can compute their autocorrelograms meaning the number of spikes of a neuron observed in a time windows centered around its own spikes.
For this we can use the function compute_autocorrelogram.
We need to specifiy the binsize and windowsize to bin the spike train.
autocorrs = nap.compute_autocorrelogram(
group=ts_group, binsize=100, windowsize=1000, time_units="ms", ep=epoch # ms
)
print(autocorrs, "\n")
Out:
0 1
-0.9 0.9475 1.05
-0.8 0.9750 1.10
-0.7 0.9725 0.97
-0.6 1.0150 1.05
-0.5 0.9425 0.85
-0.4 0.9575 0.89
-0.3 0.9425 0.79
-0.2 1.0250 1.11
-0.1 1.0750 0.96
0.0 0.0000 0.00
0.1 1.0750 0.96
0.2 1.0250 1.11
0.3 0.9425 0.79
0.4 0.9575 0.89
0.5 0.9425 0.85
0.6 1.0150 1.05
0.7 0.9725 0.97
0.8 0.9750 1.10
0.9 0.9475 1.05
The variable autocorrs is a pandas DataFrame with the center of the bins for the index and each columns is a neuron.
Similarly, we can compute crosscorrelograms meaning how many spikes of neuron 1 do I observe whenever neuron 0 fires. Here the function
is called compute_crosscorrelogram and takes a TsGroup as well.
crosscorrs = nap.compute_crosscorrelogram(
group=ts_group, binsize=100, windowsize=1000, time_units="ms" # ms
)
print(crosscorrs, "\n")
Out:
0
1
-0.9 1.040
-0.8 1.110
-0.7 1.000
-0.6 1.005
-0.5 1.090
-0.4 0.875
-0.3 0.980
-0.2 1.120
-0.1 1.055
0.0 1.045
0.1 1.010
0.2 1.040
0.3 1.070
0.4 1.045
0.5 0.985
0.6 0.965
0.7 1.055
0.8 1.170
0.9 0.980
Column name (0, 1) is read as cross-correlogram of neuron 0 and 1 with neuron 0 being the reference time.
Peri-Event Time Histogram (PETH)
A second way to examine the relationship between spiking and an event (i.e. stimulus) is to compute a PETH. pynapple uses the function compute_perievent to center spike time around the timestamps of an event within a given window.
stim = nap.Tsd(
t=np.sort(np.random.uniform(0, 1000, 50)), d=np.random.rand(50), time_units="s"
)
peth0 = nap.compute_perievent(ts1, stim, minmax=(-0.1, 0.2), time_unit="s")
print(peth0)
Out:
Index rate ref_times
------- --------- -----------
0 nan 23.9996
1 3.33333 28.0529
2 nan 46.6504
3 3.33333 72.9137
4 nan 73.5197
5 6.66667 80.8389
6 nan 117.547
7 nan 141.686
8 nan 154.962
9 nan 160.518
10 nan 190.095
11 6.66667 219.417
12 3.33333 228.274
13 nan 241.984
14 6.66667 264.217
15 3.33333 299.341
16 nan 316.922
17 nan 331.141
18 nan 406.893
19 nan 422.294
20 nan 430.157
21 3.33333 437.542
22 3.33333 464.405
23 nan 482.52
24 nan 487.688
25 6.66667 494.404
26 nan 507.024
27 3.33333 531.726
28 3.33333 556.851
29 3.33333 559.877
30 nan 571.838
31 nan 647.292
32 6.66667 709.389
33 3.33333 809.848
34 nan 810.405
35 3.33333 817.995
36 nan 866.084
37 nan 886.307
38 nan 894.487
39 3.33333 896.847
40 3.33333 897.95
41 6.66667 928.571
42 nan 929.996
43 nan 933.221
44 3.33333 933.409
45 3.33333 938.691
46 nan 962.453
47 3.33333 979.04
48 6.66667 979.768
49 3.33333 998.711
It is then easy to create a raster plot around the times of the stimulation event by calling the to_tsd function of pynapple to "flatten" the TsGroup peth0.
mkdocs_gallery_thumbnail_number = 2
plt.figure(figsize=(10, 6))
plt.subplot(211)
plt.plot(np.sum(peth0.count(0.01), 1), linewidth=3, color="red")
plt.xlim(-0.1, 0.2)
plt.ylabel("Count")
plt.axvline(0.0)
plt.subplot(212)
plt.plot(peth0.to_tsd(), "|", markersize=20, color="red", mew=4)
plt.xlabel("Time from stim (s)")
plt.ylabel("Stimulus")
plt.xlim(-0.1, 0.2)
plt.axvline(0.0)
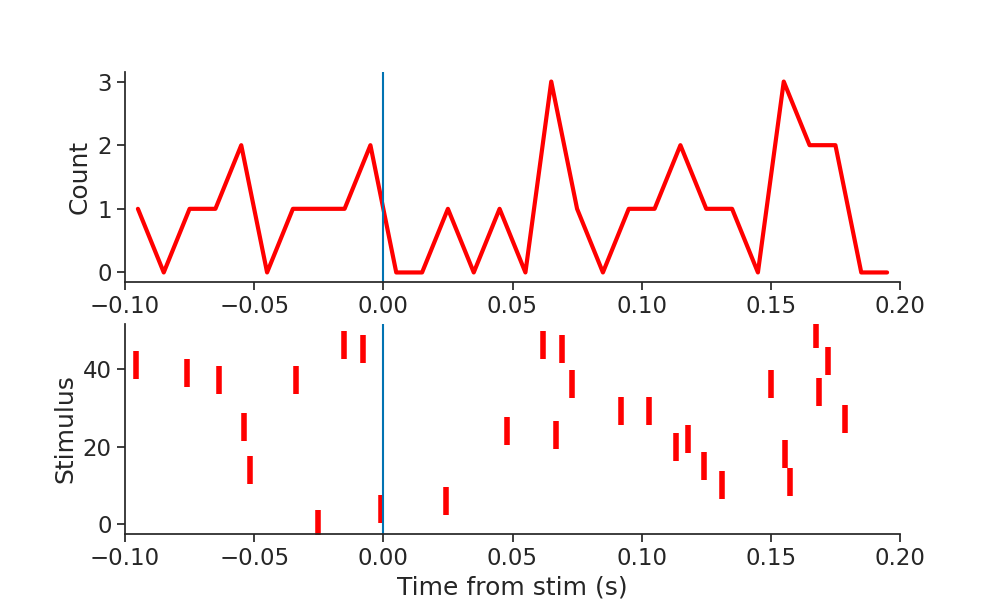
Out:
The same function can be applied to a group of neurons. In this case, it returns a dict of TsGroup
pethall = nap.compute_perievent(ts_group, stim, minmax=(-0.1, 0.2), time_unit="s")
print(pethall[1])
Out:
Index rate ref_times
------- --------- -----------
0 nan 23.9996
1 nan 28.0529
2 nan 46.6504
3 nan 72.9137
4 nan 73.5197
5 nan 80.8389
6 nan 117.547
7 3.33333 141.686
8 6.66667 154.962
9 nan 160.518
10 nan 190.095
11 3.33333 219.417
12 nan 228.274
13 nan 241.984
14 3.33333 264.217
15 nan 299.341
16 nan 316.922
17 3.33333 331.141
18 nan 406.893
19 3.33333 422.294
20 nan 430.157
21 nan 437.542
22 nan 464.405
23 nan 482.52
24 nan 487.688
25 nan 494.404
26 nan 507.024
27 nan 531.726
28 nan 556.851
29 nan 559.877
30 nan 571.838
31 nan 647.292
32 nan 709.389
33 nan 809.848
34 3.33333 810.405
35 nan 817.995
36 nan 866.084
37 nan 886.307
38 3.33333 894.487
39 nan 896.847
40 nan 897.95
41 3.33333 928.571
42 nan 929.996
43 nan 933.221
44 nan 933.409
45 nan 938.691
46 3.33333 962.453
47 3.33333 979.04
48 nan 979.768
49 3.33333 998.711
Tuning curves
pynapple can compute 1 dimension tuning curves (for example firing rate as a function of angular direction) and 2 dimension tuning curves ( for example firing rate as a function of position). In both cases, a TsGroup object can be directly passed to the function.
First we will create the 2D features:
dt = 0.1
features = np.vstack((np.cos(np.arange(0, 1000, dt)), np.sin(np.arange(0, 1000, dt)))).T
# features += np.random.randn(features.shape[0], features.shape[1])*0.05
features = nap.TsdFrame(
t=np.arange(0, 1000, dt),
d=features,
time_units="s",
time_support=epoch,
columns=["a", "b"],
)
print(features)
plt.figure(figsize=(15, 7))
plt.subplot(121)
plt.plot(features[0:100])
plt.title("Features")
plt.xlabel("Time(s)")
plt.subplot(122)
plt.title("Features")
plt.plot(features["a"][0:100], features["b"][0:100])
plt.xlabel("Feature a")
plt.ylabel("Feature b")
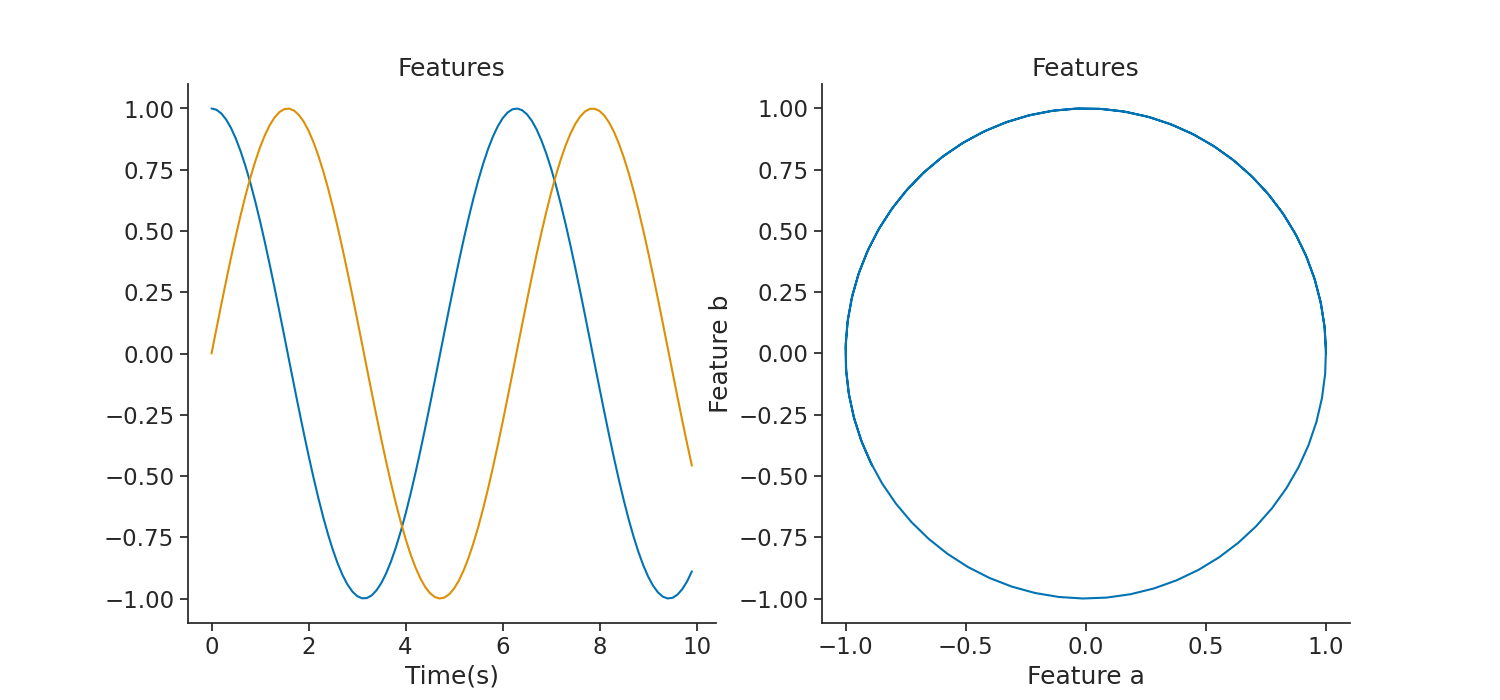
Out:
Time (s) a b
---------- -------- --------
0.0 1 0
0.1 0.995 0.09983
0.2 0.98007 0.19867
0.3 0.95534 0.29552
0.4 0.92106 0.38942
0.5 0.87758 0.47943
0.6 0.82534 0.56464
0.7 0.76484 0.64422
0.8 0.69671 0.71736
0.9 0.62161 0.78333
1.0 0.5403 0.84147
1.1 0.4536 0.89121
1.2 0.36236 0.93204
1.3 0.2675 0.96356
1.4 0.16997 0.98545
1.5 0.07074 0.99749
1.6 -0.0292 0.99957
1.7 -0.12884 0.99166
1.8 -0.2272 0.97385
1.9 -0.32329 0.9463
2.0 -0.41615 0.9093
2.1 -0.50485 0.86321
2.2 -0.5885 0.8085
2.3 -0.66628 0.74571
2.4 -0.73739 0.67546
2.5 -0.80114 0.59847
2.6 -0.85689 0.5155
2.7 -0.90407 0.42738
2.8 -0.94222 0.33499
2.9 -0.97096 0.23925
...
997.0 -0.44006 -0.89797
997.1 -0.34822 -0.93741
997.2 -0.25289 -0.96749
997.3 -0.15504 -0.98791
997.4 -0.05564 -0.99845
997.5 0.04432 -0.99902
997.6 0.14383 -0.9896
997.7 0.24191 -0.9703
997.8 0.33757 -0.9413
997.9 0.42986 -0.9029
998.0 0.51785 -0.85547
998.1 0.60066 -0.7995
998.2 0.67748 -0.73554
998.3 0.74753 -0.66423
998.4 0.81011 -0.58628
998.5 0.86459 -0.50248
998.6 0.91043 -0.41365
998.7 0.94718 -0.3207
998.8 0.97447 -0.22453
998.9 0.99201 -0.12613
999.0 0.99965 -0.02646
999.1 0.9973 0.07347
999.2 0.98498 0.17267
999.3 0.96282 0.27014
999.4 0.93104 0.36491
999.5 0.88996 0.45604
999.6 0.83999 0.54261
999.7 0.78162 0.62375
999.8 0.71544 0.69867
999.9 0.64212 0.7666
dtype: float64, shape: (10000, 2)
Text(732.5909090909089, 0.5, 'Feature b')
Here we call the function compute_2d_tuning_curves.
To check the accuracy of the tuning curves, we will display the spikes aligned to the features with the function value_from which assign to each spikes the corresponding feature value for neuron 0.
tcurves2d, binsxy = nap.compute_2d_tuning_curves(
group=ts_group, features=features, nb_bins=10
)
ts_to_features = ts_group[1].value_from(features)
plt.figure()
plt.plot(ts_to_features["a"], ts_to_features["b"], "o", color="red", markersize=4)
extents = (
np.min(features["b"]),
np.max(features["b"]),
np.min(features["a"]),
np.max(features["a"]),
)
plt.imshow(tcurves2d[1].T, origin="lower", extent=extents, cmap="viridis")
plt.title("Tuning curve unit 0 2d")
plt.xlabel("feature a")
plt.ylabel("feature b")
plt.grid(False)
plt.show()
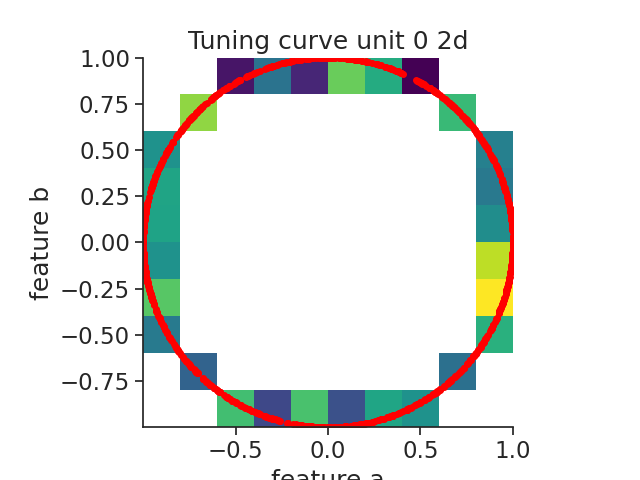
Out:
/mnt/home/gviejo/pynapple/pynapple/process/tuning_curves.py:223: RuntimeWarning: invalid value encountered in divide
count = count / occupancy
Decoding
Pynapple supports 1 dimensional and 2 dimensional bayesian decoding. The function returns the decoded feature as well as the probabilities for each timestamps.
First we generate some artificial "place fields" in 2 dimensions based on the features.
This part is just to generate units with a relationship to the features (i.e. "place fields")
times = features.as_units("us").index.values
ft = features.values
alpha = np.arctan2(ft[:, 1], ft[:, 0])
bins = np.repeat(np.linspace(-np.pi, np.pi, 13)[::, np.newaxis], 2, 1)
bins += np.array([-2 * np.pi / 24, 2 * np.pi / 24])
ts_group = {}
for i in range(12):
ts = times[(alpha >= bins[i, 0]) & (alpha <= bins[i + 1, 1])]
ts_group[i] = nap.Ts(ts, time_units="us")
ts_group = nap.TsGroup(ts_group, time_support=epoch)
print(ts_group)
Out:
Index rate
------- ------
0 1.248
1 1.665
2 1.666
3 1.664
4 1.665
5 1.67
6 1.671
7 1.673
8 1.665
9 1.666
10 1.667
11 1.248
To decode we need to compute tuning curves in 2D.
import warnings
warnings.filterwarnings("ignore")
tcurves2d, binsxy = nap.compute_2d_tuning_curves(
group=ts_group,
features=features,
nb_bins=10,
ep=epoch,
minmax=(-1.0, 1.0, -1.0, 1.0),
)
Then we plot the "place fields".
plt.figure(figsize=(20, 9))
for i in ts_group.keys():
plt.subplot(2, 6, i + 1)
plt.imshow(
tcurves2d[i], extent=(binsxy[1][0], binsxy[1][-1], binsxy[0][0], binsxy[0][-1])
)
plt.xticks()
plt.show()
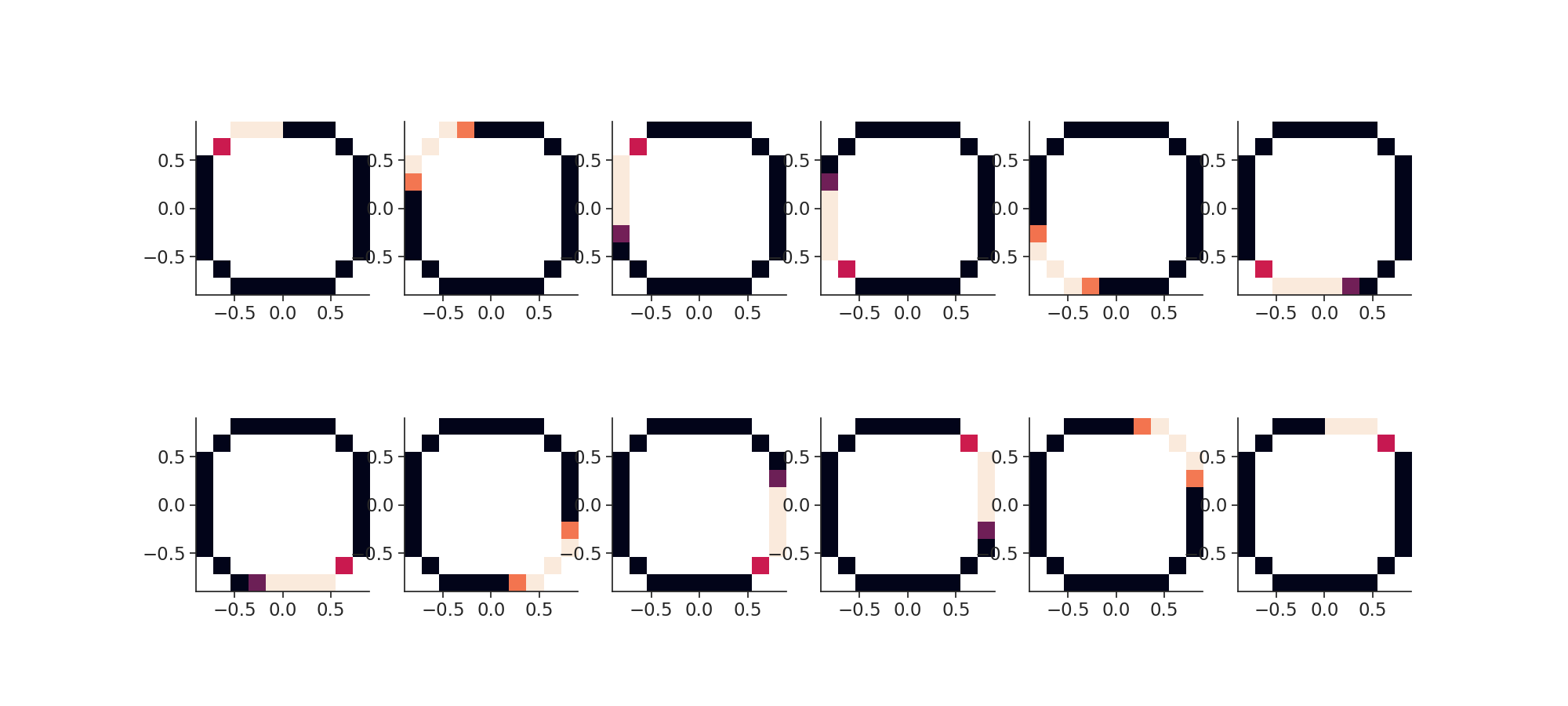
Then we call the actual decoding function in 2d.
decoded, proba_feature = nap.decode_2d(
tuning_curves=tcurves2d,
group=ts_group,
ep=epoch,
bin_size=0.1, # second
xy=binsxy,
features=features,
)
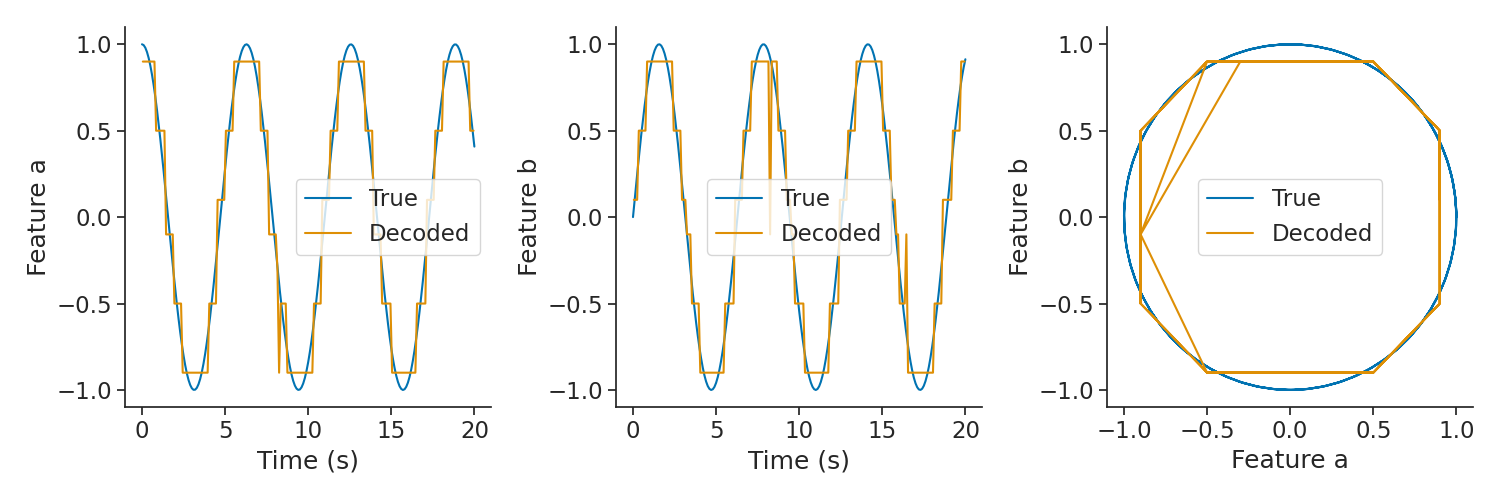
plt.figure(figsize=(15, 5))
plt.subplot(131)
plt.plot(features["a"].as_units("s").loc[0:20], label="True")
plt.plot(decoded["a"].as_units("s").loc[0:20], label="Decoded")
plt.legend()
plt.xlabel("Time (s)")
plt.ylabel("Feature a")
plt.subplot(132)
plt.plot(features["b"].as_units("s").loc[0:20], label="True")
plt.plot(decoded["b"].as_units("s").loc[0:20], label="Decoded")
plt.legend()
plt.xlabel("Time (s)")
plt.ylabel("Feature b")
plt.subplot(133)
plt.plot(
features["a"].as_units("s").loc[0:20],
features["b"].as_units("s").loc[0:20],
label="True",
)
plt.plot(
decoded["a"].as_units("s").loc[0:20],
decoded["b"].as_units("s").loc[0:20],
label="Decoded",
)
plt.xlabel("Feature a")
plt.ylabel("Feature b")
plt.legend()
plt.tight_layout()
plt.show()
Randomization
Pynapple provides some ready-to-use randomization methods to compute null distributions for statistical testing. Different methods preserve or destroy different features of the data, here's a brief overview.
shift_timestamps shifts all the timestamps in a Ts object by the same random amount, wrapping the end of the time support to its beginning. This randomization preserves the temporal structure in the data but destroys the temporal relationships with other quantities (e.g. behavioural data).
When applied on a TsGroup object, each series in the group is shifted independently.
ts = nap.Ts(t=np.sort(np.random.uniform(0, 100, 10)), time_units="ms")
rand_ts = nap.shift_timestamps(ts, min_shift=1, max_shift=20)
shuffle_ts_intervals computes the intervals between consecutive timestamps, permutes them, and generates a new set of timestamps with the permuted intervals.
This procedure preserve the distribution of intervals, but not their sequence.
ts = nap.Ts(t=np.sort(np.random.uniform(0, 100, 10)), time_units="s")
rand_ts = nap.shuffle_ts_intervals(ts)
jitter_timestamps shifts each timestamp in the data of an independent random amount. When applied with a small max_jitter, this procedure destroys the fine temporal structure of the data, while preserving structure on longer timescales.
ts = nap.Ts(t=np.sort(np.random.uniform(0, 100, 10)), time_units="s")
rand_ts = nap.jitter_timestamps(ts, max_jitter=1)
resample_timestamps uniformly re-draws the same number of timestamps in ts, in the same time support. This procedures preserve the total number of timestamps, but destroys any other feature of the original data.
ts = nap.Ts(t=np.sort(np.random.uniform(0, 100, 10)), time_units="s")
rand_ts = nap.resample_timestamps(ts)
Total running time of the script: ( 0 minutes 1.692 seconds)
Download Python source code: tutorial_pynapple_process.py